The Challenge
When the COVID-19 pandemic emerged in early 2020, there were no approved drug treatments available. Developing new drugs from scratch takes years, so we sought a faster solution - drug repurposing. By screening existing drugs for activity against SARS-CoV-2 targets, we hoped to discover candidates that could be rapidly tested in patients.
Our Approach
We focused on the viral RdRp and 3CLpro enzymes, which are essential for SARS-CoV-2 replication and transcription. Using computational methods, we:
Modeled the 3D structures of these enzyme targets.
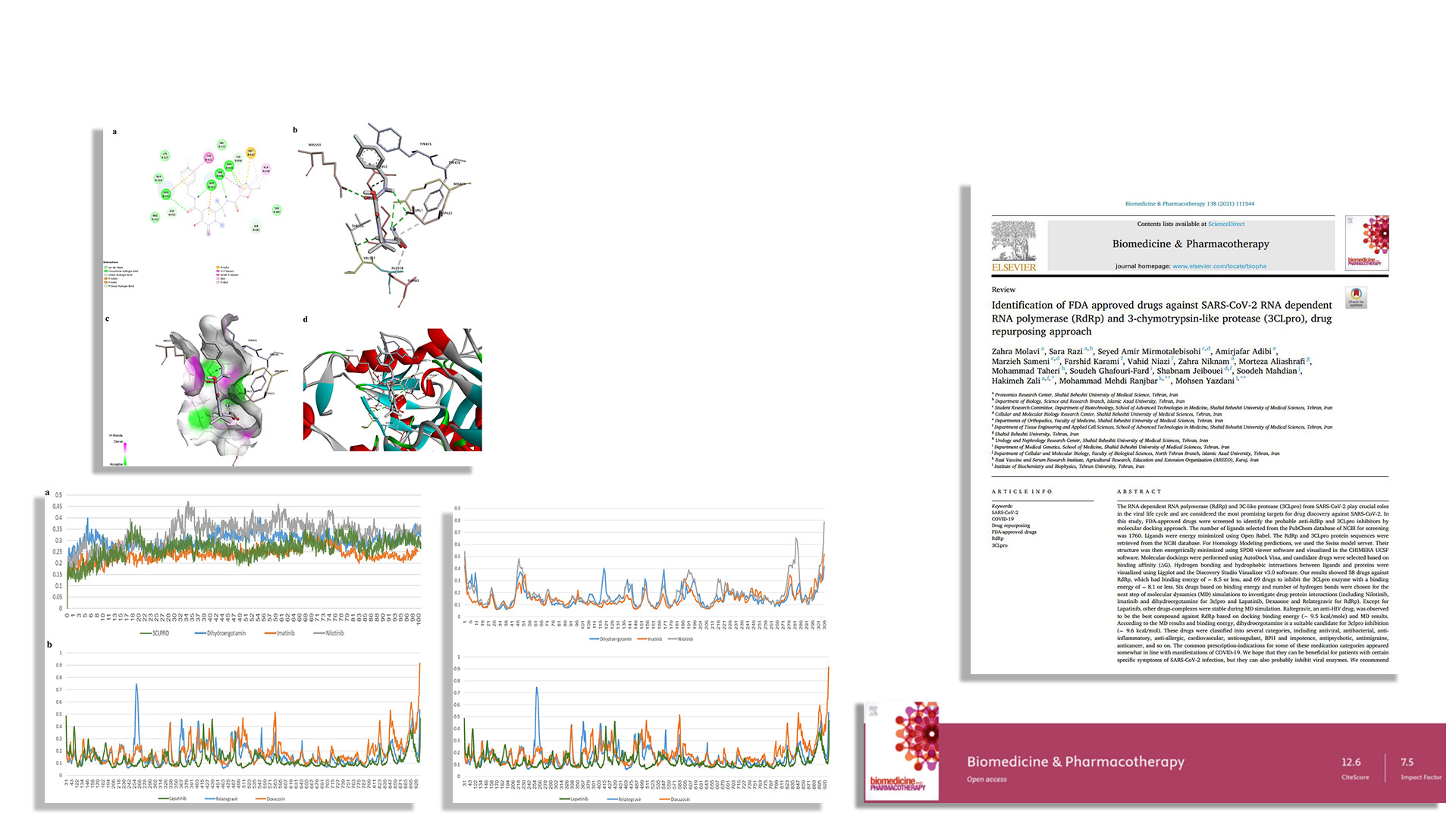
Screened 1760 FDA-approved drugs through docking simulations to identify those most likely to bind and inhibit the enzymes.
Selected top candidates based on binding energy and interactions.
Ran molecular dynamics simulations to validate stable binding over time.
Classified top candidates into drug categories relating to COVID-19 symptoms.
Our Findings
We identified 58 potential RdRp inhibitors and 69 potential 3CLpro inhibitors. The strongest binders were the antiretroviral drug raltegravir for RdRp, and anti-migraine drug dihydroergotamine for 3CLpro.
Several drug categories matched disease manifestations, including antiviral, anti-inflammatory, cardiovascular and more. This supported their potential to alleviate COVID-19 symptoms while inhibiting the virus.








